pKillerRed-CpKillerRed-C荧光表达载体质粒图谱序列抗性价格报价Biovector NTCC Inc.现货
- 价 格:¥5920
- 货 号:BiovectorpKillerRed
- 产 地:北京
- BioVector NTCC典型培养物保藏中心
- 联系人:Dr.Xu, Biovector NTCC Inc.
电话:400-800-2947 工作微信:1843439339 (QQ同号)
邮件:Biovector@163.com
手机:18901268599
地址:北京
- 已注册
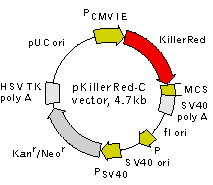
pKillerRed-C vector
cat.# FP961
The vector sequence has been compiled using the information from sequence databases, published literature, and other sources, together with partial sequences obtained by Evrogen. This vector has not been completely sequenced. | |||||||||||||||||||||||||||||||||||||||||||||||||
 Download |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| KillerRed | BspE I | Bgl II | Sac I | EcoR I | Sal I | Kpn I | Apa I | BamH I | Stops | ||||||||||||||||||||||
| Xho I | Hind III | Pst I | Sac II | Xba I# | Bcl I# | ||||||||||||||||||||||||||
| GAC.GAG.GAT. | TCC.GGA. | CTC. | AGA.T | CT | .C | GA.G | CT.C | AA.GCT.T | C | G.AAT.T | C | T.GCA. | G | TC.GAC | .GGT.A | CC. | GC | G.G | G | C.CC | G. | GG | A.TCC. | ACC.GGA. | TC | T.AG | A. | TAA. | C | TG.A | TC.A |
# – sites are blocked by dam methylation. If you wish to digest the vector with these enzymes, you will need to transform the vector into a dam- host and make fresh DNA.
Vector description
pKillerRed-C is a mammalian expression vector encoding photosensitizer KillerRed (see reporter description). The vector allows generation of fusions to the KillerRed C-terminus and expression of KillerRed fusions or KillerRed alone in eukaryotic (mammalian) cells.
KillerRed codon usage is optimized for high expression in mammalian cells (humanized) [Haas et al., 1996]. To increase mRNA translation efficiency, Kozak consensus translation initiation site is generated upstream of the KillerRed coding sequence [Kozak, 1987]. Multiple cloning site (MCS) is located between KillerRed coding sequence and SV40 polyadenylation signal (SV40 polyA).
The vector backbone contains immediate early promoter of cytomegalovirus (PCMV IE) for protein expression, SV40 origin for replication in mammalian cells expressing SV40 T-antigen, pUC origin of replication for propagation in
SV40 early promoter (PSV40) provides neomycin resistance gene (Neor) expression to select stably transfected eukaryotic cells using G418. Bacterial promoter (P) provides kanamycin resistance gene expression (Kanr) in
Generation of KillerRed fusion proteins
A localization signal or a gene of interest can be cloned into MCS of the vector. It will be expressed as a fusion to the KillerRed C-terminus when inserted in the same reading frame as KillerRed and no in-frame stop codons are present. KillerRed-tagged fusions retain fluorescent properties of the native protein allowing fusion localization in vivo. Unmodified vector will express KillerRed when transfected into eukaryotic (mammalian) cells.
Note: The plasmid DNA was isolated from dam+-methylated
Expression in mammalian cells
pKillerRed-C vector can be transfected into mammalian cells by any known transfection method. CMV promoter provides strong, constitutive expression of KillerRed or its fusions in eukaryotic cells. If required, stable transformants can be selected using G418 [Gorman, 1985].
Note: KillerRed shows no cell toxic effects before light activation. Upon green light irradiation KillerRed generates reactive oxygen species (ROS) that damage the neighboring molecules.
Propagation in
Suitable host strains for propagation in
Location of features
PCMV IE: 1-589
Enhancer region: 59-465
TATA box: 554-560
Transcription start point: 583
KillerRed
Kozak consensus translation initiation site: 606-616
Start codon (ATG): 613-615
Stop codon: 1408-1410
Last amino acid in KillerRed: 1327-1329
MCS: 1330-1417
SV40 early mRNA polyadenylation signal
Polyadenylation signals: 1550-1555 & 1579-1584
mRNA 3' ends: 1588 & 1600
f1 single-strand DNA origin: 1647-2102
Bacterial promoter for expression of Kanr gene
-35 region: 2164-2169
-10 region: 2187-2192
Transcription start point: 2199
SV40 origin of replication: 2443-2578
SV40 early promoter
Enhancer (72-bp tandem repeats): 2276-2347 & 2348-2419
21-bp repeats: 2423-2443, 2444-2464 & 2466-2486
Early promoter element: 2499-2505
Major transcription start points: 2495, 2533, 2539 & 2544
Kanamycin/neomycin resistance gene
Neomycin phosphotransferase coding sequences:
Start codon (ATG): 2627-2629
Stop codon: 3419-3421
G->A mutation to remove Pst I site: 2809
C->A (Arg to Ser) mutation to remove BssH II site: 3155
Herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signal
Polyadenylation signals: 3657-3662 & 3670-3675
pUC plasmid replication origin: 4006-4649
References:
Gorman C. High efficiency gene transfer into mammalian cells. In DNA cloning: A Practical Approach, Vol. II. Ed. D. M. Glover. (IRL Press, Oxford, U.K.). 1985; 143-90.
Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996; 6 (3):315-24. / pmid: 8805248
Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987; 15 (20):8125-48. / pmid: 3313277
Notice to Purchaser:
KillerRed-related materials (also referred to as "Products") are intended for research use only.
The CMV promoter is covered under U.S. Patents 5,168,062 and 5,385,839, and its use is permitted for research purposes only. Any other use of the CMV promoter requires a license from the University of Iowa Research Foundation, 214 Technology Innovation Center, Iowa City, IA 52242.
- 公告/新闻




