| 出品公司: | Agilent |
|---|---|
| 载体名称: | pCFB-EGSH |
| 质粒类型: | 逆病毒载体 |
| 高拷贝/低拷贝: | 低拷贝 |
| 克隆方法: | 限制性内切酶,多克隆位点 |
| 启动子: | CMV |
| 载体大小: | 6982 bp |
| 5' 测序引物及序列: | 5′-CTCTGAATACTTTCAACAAGTTAC -3′ |
| 3' 测序引物及序列: | 5′-GGCTGCCGACCCCGGGGGTGG -3′ |
| 载体标签: | 3x HA (C-端) |
| 载体抗性: | 氨苄青霉素 |
| 筛选标记: | 潮霉素(Hygromycin) |
| 克隆菌株: | DH5α 等 |
| 宿主细胞(系): | 常规细胞系(293、CV-1、CHO等) |
| 备注: | -- |
| 产品目录号: | 240028 |
| 稳定性: | 稳表达 |
| 组成型/诱导型: | 组成型 |
| 病毒/非病毒: | 逆转录病毒 |
- BioVector NTCC典型培养物保藏中心
- 联系人:Dr.Xu, Biovector NTCC Inc.
电话:400-800-2947 工作微信:1843439339 (QQ同号)
邮件:Biovector@163.com
手机:18901268599
地址:北京
- 已注册
载体基本信息
载体质粒图谱和多克隆位点信息
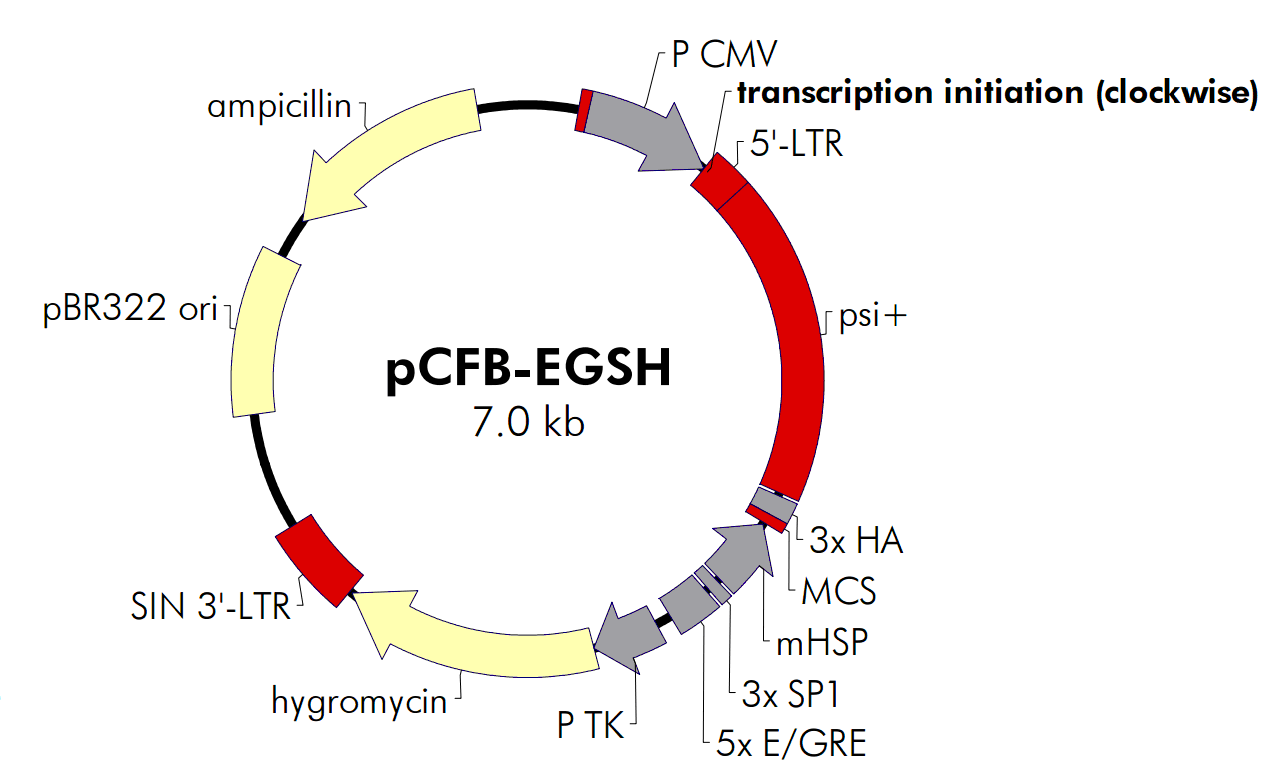

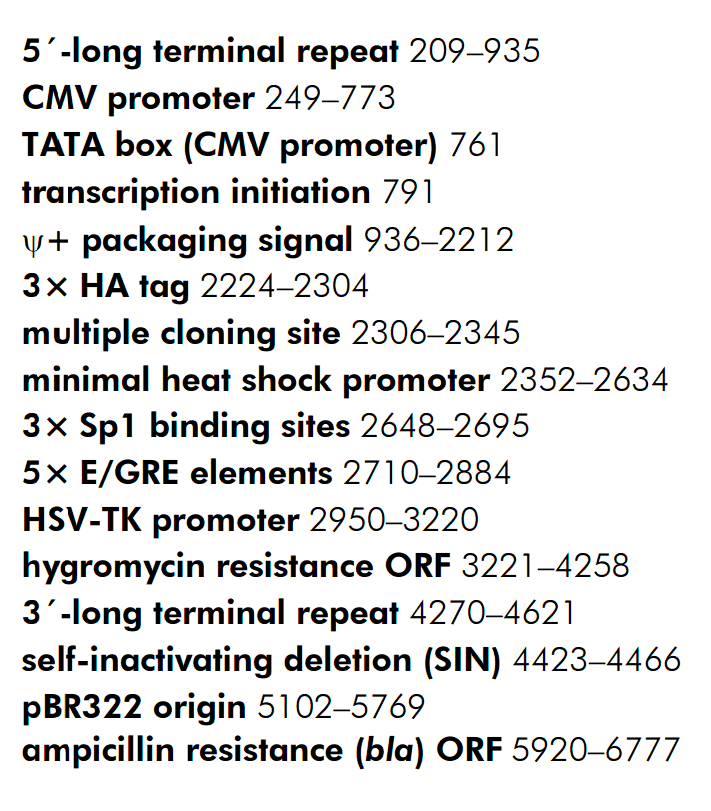
载体简介
pCFB-EGSH载体描述DNA vector-based systems that allow precise control of gene expression in vivo have become invaluable for the study of gene function in a variety of organisms, particularly when applied to the study of developmental and other biological processes for which the timing or dosage of gene expression is critical to gene function. Such systems have also been successfully used to overexpress toxic or disease-causing genes, to induce gene targeting, and to express antisense RNA. Inducible systems are currently being used by pharmaceutical companies to facilitate screening for inhibitors of clinically relevant biological pathways, and potential applications for gene therapy are being explored.The Agilent Complete Control ecdysone-inducible plasmid vectors are based on the insect molting hormone ecdysone, which can stimulate transcriptional activation in mammalian cells harboring the ecdysone receptor protein from Drosophila melanogaster.2 The system has a numberof advantages over alternative systems. Firstly, the lipophilic nature and short in vivo half-life of the ecdysone analog ponasterone A (ponA) allows efficient penetrance into all tissues including brain, resulting in rapid and potent inductions and rapid clearance. Secondly, ecdysteroids are not known, nor are they expected, to affect mammalian physiology in any measurable way. Thirdly, the heterodimeric ponA responsive receptor and receptor DNA recognition element have been genetically altered such that trans-activation of endogenous genes by the ecdysone receptor, or of the ponA-responsive expression cassette by endogenous transcription factors, is extremely unlikely. In addition, it has been found that in the absence of inducer the heterodimer remains bound at the promoter in a complex with corepressors and histone deacetylase, and is thus tightly repressed until ligand binding, at which time high-level transcriptional activation occurs (i.e., the heterodimer is converted from a tight repressor to a transactivator). In transient assays and stable cell lines harboring receptor expression plasmids in combination with a plasmid bearing an inducible luciferase expression cassette, induction ratios of 1,000-fold have been achieved.3A limitation to the use of plasmid-based vectors for controlled gene expression is the fact that many cell types of academic, industrial or clinical interest are difficult or virtually impossible to transfect using current transfection methods. In particular, primary human cell lines, lymphocytes, neurons and other nondividing cells are best transduced using viral delivery systems. The most popular and user-friendly of these are the retroviral vectors. Infection with retroviruses often yields transduction efficiencies close to 100%, and the proviral copy number can be easily controlled by varying the multiplicity of infection (MOI). This latter feature is particularly important for inducible systems, for which low basal expression and high induction ratios are affected by copy number. Thus infection of the target cell with virus at an optimal MOI should yield a high frequency of clones capable of mediating desirable expression profiles without exhaustive colony screening.With the vectors pFB-ERV and pCFB-EGSH, we have adapted the ecdysone inducible components of the Complete Control System for retroviral delivery. Used together, we have attained induction ratios of >1,000-fold with these vectors in tissue culture cells. OVERVIEW OF ECDYSONE-REGULATABLE GENE EXPRESSIONThe ecdysone receptor (EcR) is a member of the retinoid-X-receptor (RXR) family of nuclear receptors and is composed of three domains: an N-terminal activation domain (AD), a central DNA-binding domain (DBD), and a C-terminal ligand-binding and dimerization domain (LBD). In insect cells, EcR and the nuclear receptor ultraspiracle (USP) form a promoterbound heterodimer, which regulates transcription (see Figure 1). In the absence of ecdysone, the receptor heterodimer binds to corepressors and tightly represses transcription.4 When ecdysone binds to the EcR LBD, the corepressors are released, coactivators are recruited to the complex, and transcriptional activation is enabled.In mammalian cells harboring the EcR gene, EcR heterodimerizes with RXR, the mammalian homologue of USP. The EcR–RXR heterodimer binds to multiple copies of the ecdysone-responsive element (EcRE), and in the absence of ponA, represses transcription of an expression cassette. When ponA binds to the receptor, the receptor complex activates transcription of a reporter gene or a gene of interest. To avoid pleiotropic interactions with endogenous pathways in mammalian host cells, both the EcRE recognition sequence and the EcR protein were modified.The EcRE sequence was modified to create a synthetic recognition site that does not bind any endogenous transcription factors. The wild-type EcRE sequence consists of two inverted repeat sequences separated by a single nucleotide: AGTGCA N TGCACT. The EcRE sequence was changed to AGTGCA N1 TGTTCT (and renamed E/GRE). Recognition of the synthetic E/GRE recognition sequence by either a steroid receptor or a wild-type RXR heterodimer receptor is extremely unlikely, as these receptors recognized only the wild-type perfect inverted repeat. The E/GRE recognition sequence has imperfect inverted half sites separated by one nucleotide. A wild-type RXR heterodimer requires single nucleotide separation of the inverted repeats, and the majority bind to direct repeats rather than inverted repeats (EcRE is an exception).The EcR protein was modified to create a synthetic ecdysone-binding receptor that does not transactivate any host genes. Three amino acids in the EcR DBD were mutated to change its DNA-binding specificity to that of the glucocorticoid receptor (GR), which recognizes the half-site AGAACA.2 Like all steroid receptors and unlike RXR receptors, the GR protein homodimerizes and recognizes two inverted repeat sequences separated by three nucleotides. The GR–EcR fusion protein (GEcR) retains the ability to dimerize with RXR and activate, with ponA-dependence, reporter genes that contain the synthetic E/GRE recognition sequence.The GEcR receptor was further modified by replacing the EcR AD with the more potent VP16 AD. The result of all the modifications is the synthetic ecdysone-binding receptor VgEcR. VgEcR is a fusion of the ligand-binding and dimerization domain of the D. melanogaster ecdysone receptor, the DNA-binding domain of the glucocorticoid receptor, and the transcription activation domain of herpes simplex virus (HSV) VP16OVERVIEW OF REPLICATION-DEFECTIVE RETROVIRAL GENE TRANSFER SYSTEMSNon-replicating retroviral vectors contain all of the cis elements required for transcription of mRNA molecules encoding a gene of interest, and packaging of these transcripts into infectious virus particles (Figure 2). The vectors are typically comprised of an E. coli plasmid backbone containing a pair of 600 base pair viral long terminal repeats (LTRs) between which the gene of interest is inserted. The LTR is divided into 3 regions. The U3 region contains the retroviral promoter/enhancer. The U3 region is flanked in the 3′ direction by the R region, which contains the viral polyadenylation signal (pA), followed by the U5 region which, along with R, contains sequences that are critical for reverse transcription. Expression of the viral RNA is initiated within the U3 region of the 5′ LTR and is terminated in the R region of the 3′ LTR. Between the 5′ LTR and the coding sequence for the gene of interest resides an extended version of the viral packaging signal (ψ+), which is required in cis for the viral RNA to be packaged into virion particles.In order to generate infectious virus particles that carry the gene of interest, specialized packaging cell lines have been generated that contain chromosomally integrated expression cassettes for viral Gag, Pol and Env proteins, all of which are required in trans to make virus. The gag gene encodes internal structural proteins, pol encodes reverse transcriptase (RT) and integrase, and the env gene encodes the viral envelope protein, which resides on the viral surface and facilitates infection of the target cell by direct interaction with cell type-specific receptors; thus the host range of the virus is dictated not by the DNA vector but by the choice of the env gene used to construct the packaging cell. The packaging cell line is transfected with the vector DNA, and at this point either stable viral producer cell lines may be selected (providing the vector has an appropriate selectable marker), or mRNAs that are transiently transcribed from the vector are encapsidated and bud off into the cell supernatant. These supernatants are collected, and used to infect target cells. Upon infection of the target cell, the viral RNA molecule is reverse transcribed by RT (which is present in the virion particle), and the cDNA of the gene of interest, flanked by the LTRs, is integrated into the host DNA. Because the vector itself carries none of the viral proteins, once a target cell is infected the LTR expression cassette is incapable of proceeding through another round of virus production. Recent advances in transfection technology have allowed the production of high titer viral supernatants following transient cotransfection of the viral vector together with expression vectors encoding the gag, pol and env genes (Figure 2),5, 6 obviating the need for the production and maintenance of stable packaging cell lines. For example, Agilent pVPack gag-pol and env-expressing packaging vectors consistently give rise to titers of >107 infectious units (IU)/ml when cotransfected with the pFB-hrGFP control vector (Agilent Catalog #240027), using a 293-derived cell line for virus production. Description of the VectorsThe pFB-ERV vector was derived from the high-titer MoMLV vector pFBNeo5 for efficient delivery of the ecdysone receptor proteins VgEcR and RXR (Figure 3). In the vector pFB-ERV the ecdysone receptor and the neomycin-resistance open reading frame (ORF) are expressed from a tricistronic message with the neomycin resistance ORF expressed at the end of the message. Thus, maintenance of infected cell lines in G418 ensures expression of the transcript encoding the receptor genes. The tricistronic transcript is expressed from the CMV promoter, which is flanked by unique EcoR I and Fse I sites so that a cell type-specific promoter of interest may be substituted. The viral promoter within the 3′ LTR has been deleted to make this a self-inactivating (SIN) vector. Upon infection and chromosomal integration into the target cell genome, the SIN deletion is transferred to the 5′ LTR, resulting in an integrated expression cassette in which only the CMV promoter is active. Cells containing an estimated single integrated viral expression cassette can be selected in as high as 1 mg/ml G418, although 600 μg/ml is routinely used.The vector pCFB-EGSH contains an ecdysone-inducible expression cassette inserted between the viral LTRs in the antisense orientation relative to that for the viral promoter (see Figure 4). The U3 promoter within the 5′ LTR of the vector has been replaced with the CMV promoter to increase production of viral RNA in packaging cells, thereby increasing the titer of the viral supernatants. Potential interference from the proviral 5′ LTR is obviated due to the SIN deletion. The inducible expression cassette contains a multiple cloning site that contains three contiguous copies of the HA epitope(3× HA) positioned for fusion at the C-terminus of the protein of interest. A second expression cassette in which the hygromycin-resistance gene is expressed from the TK promoter is located downstream (relative to transcription from the LTRs) of the inducible cassette. A pBR322 origin and ampicillin-resistance gene allow pCFB-EGSH to be propagated in prokaryotes.The pCFB-EGSH-Luc vector contains the luciferase reporter gene and is intended for use as a positive control vector to test the expression of the VgEcR and RXR receptors in pFB-ERV-containing cell lines. The pCFB-EGSH-Luc vector is derived from the pCFB-EGSH vector and has the luciferase gene inserted in the MCS. The pCFB-EGSH-Luc vector does not contain the HA epitope sequence. pCFB-EGSH载体限制性酶切位点pCFB-EGSH, 6982 bp version 075003Enzymes with 1-10 cleavage sites: #sites -- Bp position of recognition site -- AatII 7 328, 381, 464, 650, 1153 3244, 6908 Acc65I 3 822, 2022, 4508 AccI 4 2143, 2603, 2885, 4868 AccIII 6 2258, 3442, 3979, 4115, 4228 4429 AclI 2 6215, 6588 AcuI 2 5624, 6672 AflII 4 202, 1440, 2333, 4381 AflIII 6 164, 2277, 2564, 3132, 4460 5097 AgeI 1 2016 AhdI 6 862, 908, 1449, 4548, 4594 5985 Alw44I 6 1229, 3501, 3803, 4913, 5411 6657 AlwNI 1 5508 ApaI 1 1413 ApoI 5 87, 1303, 2905, 3068, 3464 AscI 1 4452 AseI 1 6161 AsiSI 1 3581 AvaI 10 785, 818, 1416, 2194, 2647 2705, 2938, 3203, 4471, 4504 BanII 4 939, 1413, 2306, 4625 BbeI 3 790, 1831, 4476 BbsI 1 6975 BceAI 8 1174, 1574, 2147, 3498, 3928 4059, 4163, 5583 BciVI 6 831, 2162, 2273, 4517, 5306 6833 BclI 1 4423 BfrBI 2 774, 4434 BglI 4 291, 413, 484, 6104 BglII 2 1853, 2879 BlpI 2 2545, 2556 Bme1580I 8 1229, 1413, 1865, 3501, 3803 4913, 5411, 6657 BmgBI 1 2563 BmrI 3 501, 4843, 6035 BmtI 6 6, 16, 26, 197, 2346 4376 BpmI 5 1974, 3868, 3922, 4329, 6075 Bpu10I 2 1723, 2308 BpuEI 4 5203, 5465, 5742, 6610 BsaAI 4 545, 2176, 4439, 4849 BsaBI 1 2915 BsaI 9 869, 890, 957, 1589, 1977 4555, 4576, 4643, 6057 BseRI 5 893, 1243, 1736, 1775, 4579 BseYI 3 3048, 4217, 5401 BsiHKAI 8 1229, 2306, 3501, 3803, 4913 5411, 6572, 6657 BsiWI 2 2268, 2318 BsmBI 7 1151, 1268, 1512, 1571, 1757 3283, 4746 BspHI 3 5817, 6825, 6930 BspMI 1 3525 BsrBI 7 2935, 2941, 3610, 3968, 4285 5028, 6829 BsrDI 3 3211, 6044, 6226 BsrGI 1 1716 BssHII 1 4453 BssSI 5 3321, 3800, 5270, 6654, 6961 Bst1107I 1 4868 BstAPI 3 3520, 3796, 4915 BstBI 1 3071 BstEII 1 1521 BstXI 1 2920 Bsu36I 1 1451 BtgI 7 151, 567, 1106, 2962, 3573 3929, 3998 BtgZI 3 556, 2151, 3878 BtsI 2 6383, 6411 Cfr10I 3 2016, 3559, 6070 DraI 4 2339, 5854, 5873, 6565 DraIII 3 2049, 3504, 3797 DrdI 4 3719, 4100, 4786, 5199 EagI 8 1136, 2873, 2932, 2944, 3426 3591, 4161, 4263 EarI 4 1509, 1739, 4975, 6779 EciI 9 713, 1099, 1731, 1750, 2060 2652, 5169, 5315, 6143 Eco57MI 7 1974, 3868, 3922, 4329, 5624 6075, 6672 EcoICRI 1 2306 EcoNI 1 1822 EcoO109I 4 1652, 2100, 4445, 6965 EcoRI 3 2905, 3068, 3464 EcoRV 1 2915 FspI 1 6210 HaeII 6 790, 1831, 2611, 4476, 4971 5341 HincII 4 2885, 3183, 4055, 6529 HindIII 1 2330 KasI 3 790, 1831, 4476 KpnI 3 822, 2022, 4508 MluI 3 2277, 3132, 4460 MlyI 10 617, 812, 1195, 1220, 2039 4291, 4498, 4997, 5468, 5985 MmeI 8 859, 1558, 2700, 3287, 3821 4545, 5287, 5471 MscI 3 1002, 1543, 1843 MslI 8 159, 568, 2957, 3077, 4427 6238, 6397, 6756 MunI 2 11, 21 NarI 3 790, 1831, 4476 NcoI 2 567, 3573 NdeI 6 440, 1839, 1847, 2295, 3670 4919 NheI 6 6, 16, 26, 197, 2346 4376 NotI 3 2931, 2943, 4262 NsiI 2 774, 4434 NspI 5 164, 244, 3014, 4732, 5097 PacI 1 2324 PciI 2 164, 5097 PfoI 3 946, 4632, 4741 PleI 10 617, 812, 1195, 1220, 2039 4291, 4498, 4997, 5468, 5985 PmeI 1 2338 PmlI 1 2176 PpuMI 3 1652, 2100, 4445 PshAI 2 1190, 3244 PsiI 1 2892 PspOMI 1 1413 PstI 7 1353, 1535, 2505, 2910, 3161 3554, 6231 PvuI 3 1209, 3582, 6357 PvuII 1 2526 RsrII 2 3101, 3628 SacI 1 2306 SacII 2 151, 3998 SalI 1 2885 SanDI 1 4445 SapI 1 4974 ScaI 3 2695, 4189, 6468 SexAI 1 1649 SfoI 3 790, 1831, 4476 SmaI 7 818, 1416, 2194, 2647, 2705 3203, 4504 SmlI 9 202, 1440, 2333, 2938, 4381 5203, 5465, 5742, 6610 SnaBI 2 545, 4439 SpeI 1 1072 SphI 1 244 SrfI 1 1415 SspI 1 6792 StuI 1 2312 StyI 5 567, 881, 1679, 3573, 4567 TatI 9 424, 504, 537, 588, 1716 2695, 4189, 4903, 6468 TfiI 6 2482, 3317, 3439, 3639, 3890 5072 Tsp45I 8 1457, 1666, 3129, 3512, 4752 4847, 6247, 6458 TspDTI 10 34, 930, 2955, 3221, 3668 4347, 4616, 5866, 5968, 6271 TspGWI 7 1296, 2271, 2600, 3737, 3836 6440, 6757 Tth111I 6 808, 1648, 3278, 3722, 4494 4841 XbaI 1 2212 XhoI 1 2938 XhoII 9 1853, 2879, 3199, 5738, 5749 5835, 5847, 6615, 6632 XmaI 7 818, 1416, 2194, 2647, 2705 3203, 4504 XmnI 2 2385, 6585 ZraI 7 328, 381, 464, 650, 1153 3244, 6908Enzymes that do NOT cut molecule:AarI AleI BamHI BbvCI BlnI BsgI BsmI ClaI Eco47III FseI FspAI HpaI NaeI NgoMIV NruI SbfI SfiI SgrAI SwaI Van91I XcmI 载体序列
pCFB-EGSH, 6982 bp version 075003
NOTE: The following sequence has been verified for accuracy
at the junctions. The remainder of the sequence has been
obtained from existing data.
1 GAATTGCTAG CAATTGCTAG CAATTGCTAG CAATTCATAC CAGATCACCG
51 AAAACTGTCC TCCAAATGTG TCCCCCTCAC ACTCCCAAAT TCGCGGGCTT
101 CTGCCTCTTA GACCACTCTA CCCTATTCCC CACACTCACC GGAGCCAAAG
151 CCGCGGGACA TATACATGTG AAAGACCCCA CCTGTAGGTT TGGCAAGCTA
201 GCTTAAGTAA CGCCATTTTG CAAGGCATGG AAAAATACAT AACGCATGCC
251 CCATATATGG AGTTCCGCGT TACATAACTT ACGGTAAATG GCCCGCCTGG
301 CTGACCGCCC AACGACCCCC GCCCATTGAC GTCAATAATG ACGTATGTTC
351 CCATAGTAAC GCCAATAGGG ACTTTCCATT GACGTCAATG GGTGGAGTAT
401 TTACGGTAAA CTGCCCACTT GGCAGTACAT CAAGTGTATC ATATGCCAAG
451 TACGCCCCCT ATTGACGTCA ATGACGGTAA ATGGCCCGCC TGGCATTATG
501 CCCAGTACAT GACCTTATGG GACTTTCCTA CTTGGCAGTA CATCTACGTA
551 TTAGTCATCG CTATTACCAT GGTGATGCGG TTTTGGCAGT ACATCAATGG
601 GCGTGGATAG CGGTTTGACT CACGGGGATT TCCAAGTCTC CACCCCATTG
651 ACGTCAATGG GAGTTTGTTT TGGCACCAAA ATCAACGGGA CTTTCCAAAA
701 TGTCGTAACA ACTCCGCCCC ATTGACGCAA ATGGGCGGTA GGCGTGTACG
751 GTGGGAGGTC TATATAAGCA GAGATGCATC CTCACTCGGG GCGCCAGTCC
801 TCCGATTGAC TGAGTCGCCC GGGTACCCGT GTATCCAATA AACCCTCTTG
851 CAGTTGCATC CGACTTGTGG TCTCGCTGTT CCTTGGGAGG GTCTCCTCTG
901 AGTGATTGAC TACCCGTCAG CGGGGGTCTT TCATTTGGGG GCTCGTCCGG
951 GATCGGGAGA CCCCTGCCCA GGGACCACCG ACCCACCACC GGGAGGTAAG
1001 CTGGCCAGCA ACTTATCTGT GTCTGTCCGA TTGTCTAGTG TCTATGACTG
1051 ATTTTATGCG CCTGCGTCGG TACTAGTTAG CTAACTAGCT CTGTATCTGG
1101 CGGACCCGTG GTGGAACTGA CGAGTTCGGA ACACCCGGCC GCAACCCTGG
1151 GAGACGTCCC AGGGACTTCG GGGGCCGTTT TTGTGGCCCG ACCTGAGTCC
1201 AAAAATCCCG ATCGTTTTGG ACTCTTTGGT GCACCCCCCT TAGAGGAGGG
1251 ATATGTGGTT CTGGTAGGAG ACGAGAACCT AAAACAGTTC CCGCCTCCGT
1301 CTGAATTTTT GCTTTCGGTT TGGGACCGAA GCCGCGCCGC GCGTCTTGTC
1351 TGCTGCAGCA TCGTTCTGTG TTGTCTCTGT CTGACTGTGT TTCTGTATTT
1401 GTCTGAAAAT ATGGGCCCGG GCCAGACTGT TACCACTCCC TTAAGTTTGA
1451 CCTTAGGTCA CTGGAAAGAT GTCGAGCAGA TCGCTCACAA CCAGTCGGTA
1501 GATGTCAAGA AGAGACGTTG GGTTACCTTC TGCTCTGCAG AATGGCCAAC
1551 CTTTAACGTC GGATGGCCGC GAGACGGCAC CTTTAACCGA GACCTCATCA
1601 CCCAGGTTAA GATCAAGGTC TTTTCACCTG GCCCGCATGG ACACCCAGAC
1651 CAGGTCCCCT ACATCGTGAC CTGGGAAGCC TTGGCTTTTG ACCCCCCTCC
1701 CTGGGTCAAG CCCTTTGTAC ACCCTAAGCC TCCGCCTCCT CTTCCTCCAT
1751 CCGCCCCGTC TCTCCCCCTT GAACCTCCTC GTTCGACCCC GCCTCGATCC
1801 TCCCTTTATC CAGCCCTCAC TCCTTCTCTA GGCGCCCCCA TATGGCCATA
1851 TGAGATCTTA TATGGGGCAC CCCCGCCCCT TGTAAACTTC CCTGACCCTG
1901 ACATGACAAG AGTTACTAAC AGCCCCTCTC TCCAAGCTCA CTTACAGGCT
1951 CTCTACTTAG TCCAGCACGA AGTCTGGAGA CCTCTGGCGG CAGCCTACCA
2001 AGAACAACTG GACCGACCGG TGGTACCTCA CCCTTACCGA GTCGGCGACA
2051 CAGTGTGGGT CCGCCGACAC CAGACTAAGA ACCTAGAACC TCGCTGGAAA
2101 GGACCTTACA CAGTCCTGCT GACCACCCCC ACCGCCCTCA AAGTAGACGG
2151 CATCGCAGCT TGGATACACG CCGCCCACGT GAAGGCTGCC GACCCCGGGG
2201 GTGGACCATC CTCTAGACTA TTAAGCGTAG TCAGGTACGT CGTAAGGGTA
2251 AGCGTAATCC GGAACGTCGT ACGGATACGC GTAGTCTGGA ACGTCATATG
2301 GGTACGAGCT CAGGCCTCGT ACGTTAATTA AGCTTAAGTT TAAACGCTAG
2351 CTGTGTGTGA GTTCTTCTTT CTCGGTAACT TGTTGAAAGT ATTCAGAGTT
2401 CTCGTCTTGT ATTCAATAAT TACTTCTTGG CAGATTTCAG TAGTTGCAGT
2451 TGATTTACTT GGTTGCTGGT TACTTTTAAT TGATTCACTT TAACTTGCAC
2501 TTTACTGCAG ATTGTTTAGC TTGTTCAGCT GCGCTTGTTT ATTTGCTTAG
2551 CTTTCGCTTA GCGACGTGTT CACTTTGCTT GTTTGAATTG AATTGTCGCT
2601 CCGTAGACGA AGCGCCTCTA TTTATACTCC GGCGGTCGAG GGTACTCCCG
2651 GGGCGGAGCT ATGCGGGGCC GGGGCTAATC GCTAGGGGCG GGGCAGTACT
2701 CCGACCCGGG TACTGAGCTT TCAGCAAGAG AACAATGCAC TTGTCCATCG
2751 AGCTTTCAGC AAGAGAACAA TGCACTTGTC CATCGAGCTT TCAGCAAGAG
2801 AACAATGCAC TTGTCCATCG AGCTAACGAG AACAATGCAC TTAGCGGTAT
2851 CGAGAACAAT GCACTTAATA TGCGGCCGAG ATCTGTCGAC TTTATAATAA
2901 GGGCGAATTC TGCAGATATC CATCACACTG GCGGCCGCTC GAGCGGCCGC
2951 CTGCTTCATC CCCGTGGCCC GTTGCTCGCG TTTGCTGGCG GTGTCCCCGG
3001 AAGAAATATA TTTGCATGTC TTTAGTTCTA TGATGACACA AACCCCGCCC
3051 AGCGTCTTGT CATTGGCGAA TTCGAACACG CAGATGCAGT CGGGGCGGCG
3101 CGGTCCGAGG TCCACTTCGC ATATTAAGGT GACGCGTGTG GCCTCGAACA
3151 CCGAGCGACC CTGCAGCGAC CCGCTTAACA GCGTCAACAG CGTGCCGCAG
3201 ATCCCGGGGG GCAATGAGAT ATGAAAAAGC CTGAACTCAC CGCGACGTCT
3251 GTCGAGAAGT TTCTGATCGA AAAGTTCGAC AGCGTCTCCG ACCTGATGCA
3301 GCTCTCGGAG GGCGAAGAAT CTCGTGCTTT CAGCTTCGAT GTAGGAGGGC
3351 GTGGATATGT CCTGCGGGTA AATAGCTGCG CCGATGGTTT CTACAAAGAT
3401 CGTTATGTTT ATCGGCACTT TGCATCGGCC GCGCTCCCGA TTCCGGAAGT
3451 GCTTGACATT GGGGAATTCA GCGAGAGCCT GACCTATTGC ATCTCCCGCC
3501 GTGCACAGGG TGTCACGTTG CAAGACCTGC CTGAAACCGA ACTGCCCGCT
3551 GTTCTGCAGC CGGTCGCGGA GGCCATGGAT GCGATCGCTG CGGCCGATCT
3601 TAGCCAGACG AGCGGGTTCG GCCCATTCGG ACCGCAAGGA ATCGGTCAAT
3651 ACACTACATG GCGTGATTTC ATATGCGCGA TTGCTGATCC CCATGTGTAT
3701 CACTGGCAAA CTGTGATGGA CGACACCGTC AGTGCGTCCG TCGCGCAGGC
3751 TCTCGATGAG CTGATGCTTT GGGCCGAGGA CTGCCCCGAA GTCCGGCACC
3801 TCGTGCACGC GGATTTCGGC TCCAACAATG TCCTGACGGA CAATGGCCGC
3851 ATAACAGCGG TCATTGACTG GAGCGAGGCG ATGTTCGGGG ATTCCCAATA
3901 CGAGGTCGCC AACATCTTCT TCTGGAGGCC GTGGTTGGCT TGTATGGAGC
3951 AGCAGACGCG CTACTTCGAG CGGAGGCATC CGGAGCTTGC AGGATCGCCG
4001 CGGCTCCGGG CGTATATGCT CCGCATTGGT CTTGACCAAC TCTATCAGAG
4051 CTTGGTTGAC GGCAATTTCG ATGATGCAGC TTGGGCGCAG GGTCGATGCG
4101 ACGCAATCGT CCGATCCGGA GCCGGGACTG TCGGGCGTAC ACAAATCGCC
4151 CGCAGAAGCG CGGCCGTCTG GACCGATGGC TGTGTAGAAG TACTCGCCGA
4201 TAGTGGAAAC CGACGCCCCA GCACTCGTCC GGATCGGGAG ATGGGGGAGG
4251 CTAACTGATA AGCGGCCGCG ATCCGAGTTC TTCTGAGCGG GACTCTGGGG
4301 TTCGATAAAA TAAAAGATTT TATTTAGTCT CCAGAAAAAG GGGGGAATGA
4351 AAGACCCCAC CTGTAGGTTT GGCAAGCTAG CTTAAGTAAC GCCATTTTGC
4401 AAGGCATGGA AAAATACATA ACTGATCATC CGGATGCATA CGTAGGGACC
4451 CGGCGCGCCA CGCGTCCTCA CTCGGGGCGC CAGTCCTCCG ATTGACTGAG
4501 TCGCCCGGGT ACCCGTGTAT CCAATAAACC CTCTTGCAGT TGCATCCGAC
4551 TTGTGGTCTC GCTGTTCCTT GGGAGGGTCT CCTCTGAGTG ATTGACTACC
4601 CGTCAGCGGG GGTCTTTCAT TTGGGGGCTC GTCCGGGATC GGGAGACCCC
4651 TGCCCAGGGA CCACCGACCC ACCACCGGGA GGTAAGCTGG CTGCCTCGCG
4701 CGTTTCGGTG ATGACGGTGA AAACCTCTGA CACATGCAGC TCCCGGAGAC
4751 GGTCACAGCT TGTCTGTAAG CGGATGCCGG GAGCAGACAA GCCCGTCAGG
4801 GCGCGTCAGC GGGTGTTGGC GGGTGTCGGG GCGCAGCCAT GACCCAGTCA
4851 CGTAGCGATA GCGGAGTGTA TACTGGCTTA ACTATGCGGC ATCAGAGCAG
4901 ATTGTACTGA GAGTGCACCA TATGCGGTGT GAAATACCGC ACAGATGCGT
4951 AAGGAGAAAA TACCGCATCA GGCGCTCTTC CGCTTCCTCG CTCACTGACT
5001 CGCTGCGCTC GGTCGTTCGG CTGCGGCGAG CGGTATCAGC TCACTCAAAG
5051 GCGGTAATAC GGTTATCCAC AGAATCAGGG GATAACGCAG GAAAGAACAT
5101 GTGAGCAAAA GGCCAGCAAA AGGCCAGGAA CCGTAAAAAG GCCGCGTTGC
5151 TGGCGTTTTT CCATAGGCTC CGCCCCCCTG ACGAGCATCA CAAAAATCGA
5201 CGCTCAAGTC AGAGGTGGCG AAACCCGACA GGACTATAAA GATACCAGGC
5251 GTTTCCCCCT GGAAGCTCCC TCGTGCGCTC TCCTGTTCCG ACCCTGCCGC
5301 TTACCGGATA CCTGTCCGCC TTTCTCCCTT CGGGAAGCGT GGCGCTTTCT
5351 CATAGCTCAC GCTGTAGGTA TCTCAGTTCG GTGTAGGTCG TTCGCTCCAA
5401 GCTGGGCTGT GTGCACGAAC CCCCCGTTCA GCCCGACCGC TGCGCCTTAT
5451 CCGGTAACTA TCGTCTTGAG TCCAACCCGG TAAGACACGA CTTATCGCCA
5501 CTGGCAGCAG CCACTGGTAA CAGGATTAGC AGAGCGAGGT ATGTAGGCGG
5551 TGCTACAGAG TTCTTGAAGT GGTGGCCTAA CTACGGCTAC ACTAGAAGGA
5601 CAGTATTTGG TATCTGCGCT CTGCTGAAGC CAGTTACCTT CGGAAAAAGA
5651 GTTGGTAGCT CTTGATCCGG CAAACAAACC ACCGCTGGTA GCGGTGGTTT
5701 TTTTGTTTGC AAGCAGCAGA TTACGCGCAG AAAAAAAGGA TCTCAAGAAG
5751 ATCCTTTGAT CTTTTCTACG GGGTCTGACG CTCAGTGGAA CGAAAACTCA
5801 CGTTAAGGGA TTTTGGTCAT GAGATTATCA AAAAGGATCT TCACCTAGAT
5851 CCTTTTAAAT TAAAAATGAA GTTTTAAATC AATCTAAAGT ATATATGAGT
5901 AAACTTGGTC TGACAGTTAC CAATGCTTAA TCAGTGAGGC ACCTATCTCA
5951 GCGATCTGTC TATTTCGTTC ATCCATAGTT GCCTGACTCC CCGTCGTGTA
6001 GATAACTACG ATACGGGAGG GCTTACCATC TGGCCCCAGT GCTGCAATGA
6051 TACCGCGAGA CCCACGCTCA CCGGCTCCAG ATTTATCAGC AATAAACCAG
6101 CCAGCCGGAA GGGCCGAGCG CAGAAGTGGT CCTGCAACTT TATCCGCCTC
6151 CATCCAGTCT ATTAATTGTT GCCGGGAAGC TAGAGTAAGT AGTTCGCCAG
6201 TTAATAGTTT GCGCAACGTT GTTGCCATTG CTGCAGGCAT CGTGGTGTCA
6251 CGCTCGTCGT TTGGTATGGC TTCATTCAGC TCCGGTTCCC AACGATCAAG
6301 GCGAGTTACA TGATCCCCCA TGTTGTGCAA AAAAGCGGTT AGCTCCTTCG
6351 GTCCTCCGAT CGTTGTCAGA AGTAAGTTGG CCGCAGTGTT ATCACTCATG
6401 GTTATGGCAG CACTGCATAA TTCTCTTACT GTCATGCCAT CCGTAAGATG
6451 CTTTTCTGTG ACTGGTGAGT ACTCAACCAA GTCATTCTGA GAATAGTGTA
6501 TGCGGCGACC GAGTTGCTCT TGCCCGGCGT CAACACGGGA TAATACCGCG
6551 CCACATAGCA GAACTTTAAA AGTGCTCATC ATTGGAAAAC GTTCTTCGGG
6601 GCGAAAACTC TCAAGGATCT TACCGCTGTT GAGATCCAGT TCGATGTAAC
6651 CCACTCGTGC ACCCAACTGA TCTTCAGCAT CTTTTACTTT CACCAGCGTT
6701 TCTGGGTGAG CAAAAACAGG AAGGCAAAAT GCCGCAAAAA AGGGAATAAG
6751 GGCGACACGG AAATGTTGAA TACTCATACT CTTCCTTTTT CAATATTATT
6801 GAAGCATTTA TCAGGGTTAT TGTCTCATGA GCGGATACAT ATTTGAATGT
6851 ATTTAGAAAA ATAAACAAAT AGGGGTTCCG CGCACATTTC CCCGAAAAGT
6901 GCCACCTGAC GTCTAAGAAA CCATTATTAT CATGACATTA ACCTATAAAA
6951 ATAGGCGTAT CACGAGGCCC TTTCGTCTTC AA您正在向 biovector.net 发送关于产品 pCFB-EGSH逆转录病毒载体质粒-BioVector NTCC保藏中心 的询问
- 公告/新闻




