L4440-Gateway质粒线虫RNAi表达载体gateway技术兼容-BioVector NTCC保藏中心
- 价 格:¥5920
- 货 号:L4440-Gateway质粒
- 产 地:北京
- BioVector NTCC典型培养物保藏中心
- 联系人:Dr.Xu, Biovector NTCC Inc.
电话:400-800-2947 工作微信:1843439339 (QQ同号)
邮件:Biovector@163.com
手机:18901268599
地址:北京
- 已注册
L4440-Gateway质粒
Gateway-modified RNAi feeding vector L4440. Use this vector for Gateway cloning of an RNAi target sequence. A Gateway recombinational cassette was placed into the EcoRV site of vector L4440 to generate this modified plasmid. To facilitate subcloning of a given DNA insert into this plasmid, Gateway recombinational attachment sites should be incorporated into primers used for amplificiation, as outlined in the Invitrogen Gateway manual. (Gateway is a registered trademark of Invitrogen Corporation.)
This plasmid is intended for use exclusively as a teaching resource as part of the Integrated Genomics Discovery-Based Laboratory Course.
L4440
Map图谱
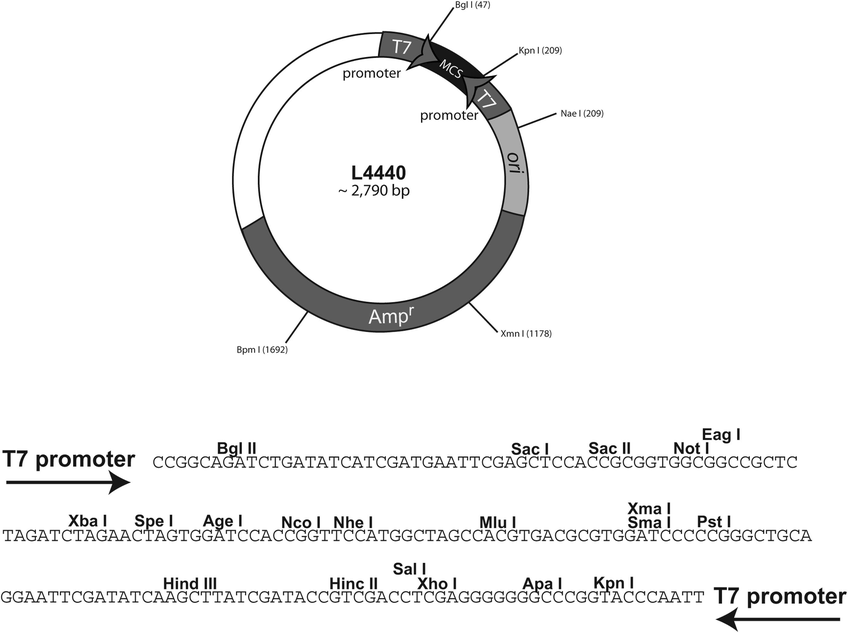
Sequence序列
AACCTGGCTTATCGAAATTAATACGACTCACTATAGGGAGACCGGCAGATCTGATATCATCGATGAATTC
GAGCTCCACCGCGGTGGCGGCCGCTCTAGAACTAGTggatccaccggttccatggctagccacgtgacgc
gtggatccCCCGGGCTGCAGGAATTCGATATCAAGCTTATCGATACCGTCGACCTCGAGGGGGGGCCCGG
TACCCAATTCGCCCTATAGTGAGTCGTATTACGCGCGCTCACTGGCCGTCGTTTTACAACGTCGTGACTG
GGAAAACCCTGGCGTTACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGC
GAAGAGGCCCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAATGGGACGCGCCCTGTA
GCGGCGCATTAAGCGCGGCGGGTGTGGTGGTTACGCGCAGCGTGACCGCTACACTTGCCAGCGCCCTAGC
GCCCGCTCCTTTCGCTTTCTTCCCTTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAAT
CGGGGGCTCCCTTTAGGGTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTAGGGTG
ATGGTTCACGTAGTGGGCCATCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTT
TAATAGTGGACTCTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGTCTATTCTTTTGATTTATAA
GGGATTTTGCCGATTTCGGCCTATTGGTTAAAAAATGAGCTGATTTAACAAAAATTTAACGCGAATTTTA
ACAAAATATTAACGCTTACAATTTAGGTGGCACTTTTCGGGGAAATGTGCGCGGAACCCCTATTTGTTTA
TTTTTCTAAATACATTCAAATATGTATCCGCTCATGAGACAATAACCCTGATAAATGCTTCAATAATATT
GAAAAAGGAAGAGTATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCCTTTTTTGCGGCATTTTGCCT
TCCTGTTTTTGCTCACCCAGAAACGCTGGTGAAAGTAAAAGATGCTGAAGATCAGTTGGGTGCACGAGTG
GGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAGAGTTTTCGCCCCGAAGAACGTTTTCCAA
TGATGAGCACTTTTAAAGTTCTGCTATGTGGCGCGGTATTATCCCGTATTGACGCCGGGCAAGAGCAACT
CGGTCGCCGCATACACTATTCTCAGAATGACTTGGTTGAGTACTCACCAGTCACAGAAAAGCATCTTACG
GATGGCATGACAGTAAGAGAATTATGCAGTGCTGCCATAACCATGAGTGATAACACTGCGGCCAACTTAC
TTCTGACAACGATCGGAGGACCGAAGGAGCTAACCGCTTTTTTGCACAACATGGGGGATCATGTAACTCG
CCTTGATCGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGCGTGACACCACGATGCCTGTA
GCAATGGCAACAACGTTGCGCAAACTATTAACTGGCGAACTACTTACTCTAGCTTCCCGGCAACAATTAA
TAGACTGGATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGCTCGGCCCTTCCGGCTGGCTGGTTTAT
TGCTGATAAATCTGGAGCCGGTGAGCGTGGGTCTCGCGGTATCATTGCAGCACTGGGGCCAGATGGTAAG
CCCTCCCGTATCGTAGTTATCTACACGACGGGGAGTCAGGCAACTATGGATGAACGAAATAGACAGATCG
CTGAGATAGGTGCCTCACTGATTAAGCATTGGTAACTGTCAGACCAAGTTTACTCATATATACTTTAGAT
TGATTTAAAACTTCATTTTTAATTTAAAAGGATCTAGGTGAAGATCCTTTTTGATAATCTCATGACCAAA
ATCCCTTAACGTGAGTTTTCGTTCCACTGAGCGTCAGACCCCGTAGAAAAGATCAAAGGATCTTCTTGAG
ATCCTTTTTTTCTGCGCGTAATCTGCTGCTTGCAAACAAAAAAACCACCGCTACCAGCGGTGGTTTGTTT
GCCGGATCAAGAGCTACCAACTCTTTTTCCGAAGGTAACTGGCTTCAGCAGAGCGCAGATACCAAATACT
GTCCTTCTAGTGTAGCCGTAGTTAGGCCACCACTTCAAGAACTCTGTAGCACCGCCTACATACCTCGCTC
TGCTAATCCTGTTACCAGTGGCTGCTGCCAGTGGCGATAAGTCGTGTCTTACCGGGTTGGACTCAAGACG
ATAGTTACCGGATAAGGCGCAGCGGTCGGGCTGAACGGGGGGTTCGTGCACACAGCCCAGCTTGGAGCGA
ACGACCTACACCGAACTGAGATACCTACAGCGTGAGCTATGAGAAAGCGCCACGCTTCCCGAAGGGAGAA
AGGCGGACAGGTATCCGGTAAGCGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGGGAAA
CGCCTGGTATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGCTCG
TCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTGGC
CTTTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGTG
AGCTGATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGAAGC
Bacteria-mediated RNAi--General outline:
1) Place gene of interest between T7 promoters in “double T7” plamsid. (slightly more difficult alternative: Place gene of
interest in hairpin/inverted repeat configuration behind T7 promoter in pBlueScript.)
note: this (and other standard cloning) should be performed in DH5 bacteria or other standard cloning strain--NOT
HT115(DE3) cells.
another note: The double T7 promoter-containing plasmid as well as control plasmids for use in feeding experiments
are availale in the 1999 FireLab vector kit. Information and kit request forms can be accessed through the Carnegie
Web site: http://www.ciwemb.edu/
2) Transform plasmid into competent HT115(DE3) bacterial cells and plate onto standard LB+ tetracycline+antibiotic plates.
(Easy competent cell and transformation protocols below)
note: the HT115 cells were a generous gift from D. Court (NCI).
another note: the HT115(DE3) strain is tetracycline resistant; nonetheless, care must be taken not to contaminate your
bacterial stock. First plate the cells onto TET plates, immediately freeze an aliquot upon receipt, and also freeze any
transformed strains in order to have reliable backup. (see freezing protocol below). In addition, the only reliable way
to verify the presence of the DE3 lysogen is by PCR, since T7 phage will not grow in the RNAseIII- background of
this cell.
yet another note: The HT115(DE3) strain is now available from the CGC. (http://biosci.cbs.umn.edu/CGC/
CGChomepage.htm)
3) Grow up culture from single colony on plates and induce expression of dsRNA using IPTG (induction protocol below).
4) Seed NGM plates with the induced culture. The culture can be used as is (for small plates containing small numbers of
hand-picked worms, eg), or the cells can be concentrated by centrifugation and spotted onto plates (for large plates containing
chunked worms, eg). The ratio of bacteria to worms is important--If the plates starve out, RNAi will not be effective. In
addition, the bacterial lawn should not be allowed to continue to grow. Cells that do grow on plates after induction are
generally cells that have lost the plasmid, cells that have lost the ability to produce T7 polymerase, or cells that are
contaminants. The inclusion of tetracycline in the plates significantly improves the results (the addition of ampicillin also helps
—in the case of amp resistant plasmids) (50ug/ml AMP,12.5ug/ml TET). IPTG included in the plates does not significantly
improve the RNAi phenotypes in my hands, but I usually include it in the NGM plates anyway, especially if the seeded plates
are not going to be used immediately (0.4mM IPTG).
5) Add worms to plate and incubate at appropriate temperature. Worms can be added by hand-picking or by adding chunks
onto wet, freshly seeded plates, or onto plates that have been allowed to dry after seeding. I generally use freshly seeded plates
in my experiments. Older seeded plates containing IPTG can also induce RNAi phenotypes with good success. We observe
phenotypes at all temperatures from 16-25C, although the expressivity and penetrance of the phenotype can vary depending
upon the incubation temperature and gene. Worms grown on dsgfp-plates have a stronger RNAi phenotypes when the plates
are incubated at lower temps (16C or 20C), with ds unc-22, phenotypes more convincing at higher temps (25C). It can take
three days before an RNAi phenotype is observed. Results vary depending on the dsRNA and the worm strains used. Freshly
seeded plates vs older seeded plates is also a consideration.
Quick procedure for making competent bacterial cells using CaCl2:
1) Inoculate overnight culture in LB + antibiotic (TET for HT115(DE3) strain + antibiotic appropriate for any plasmids in
cells) (2-5ml). Shake overnight at 37C.2) Inoculate 25 ml LB + antibiotic with overnight culture, 1:100 dilution. Grow cells to OD595= 0.4. Can grow cells in 50
ml sterile centrifuge tube.
3) Spin cells 10 min 3000 rpm at 4C.
4) Resuspend pellet in 0.5X original volume cold, sterile 50 mM CaCl2 (12.5ml). Resuspend by GENTLY pipetting up and
down a few times with a wide bore pipet--no vortexing.
5) Incubate on ice 30 min.
6) Spin as before at 4C.
7) Resuspend pellet as before in 0.1X original volume CaCl2 (2.5ml). Keep cells cold (4C).
8) Use 50-200 ul for transformation.
Cells can be used as is for up to three days (stored at 4C). The cells can be frozen by: adding glycerol to final concentration
of 10%, rapid freezing on dry ice/etoh, and storing at –80.
Transformation of CaCl2 competent HT115(DE3):
1) Add 50-200 ul competent cells to cold, sterile, polypropylene tube on ice.
2) Add 1 uL plasmid (mini-prep quality is OK) 1-100ng.
3) Incubate on ice/water bath for 30 min.
4) Immerse tube in 37C water bath for 1 min.
5) Incubate tube on ice/water bath for 2 min.
6) Add 1ml sterile SOC media. Incubate 37C with shaking for 1 hour.
7) Plate 10uL, 100uL, 250uL, and remaining culture onto 4 LB + tetracycline + other antibiotic? (plasmid resistance) plates.
Incubate 37C overnight. (These cells grow slowly, allow 36 hours for colony formation.)
Freezing bacterial stocks:
1) Inoculate fresh single colony of bacteria into 2.5 ml LB+ antibiotic(s). Grow to early stationary phase.
2) Pipette 0.25 ml 80% glycerol (sterile) and 0.75 ml culture into a sterile screw-cap freezer tube. Mix.
3) Quick freeze on dry ice/ethanol and store at -80C.
Induction of dsRNA in HT115(DE3) cells + T7 promoter containing plasmid:
1) Inoculate overnight culture of HT115(DE3) + plasmid in LB+antibiotics. Incubate 37C with shaking overnight.
(75-100ug/ml ampicillin for amp-resistant plasmids and 12.5 ug/ml tetracycline.)
2) Dilute culture 1:100 in 2xYT + antibiotics and grow to OD595=0.4. (A 25ml culture is usually enough for a small
experiment ~ 20 small plates.)
3) Induce by adding sterile IPTG to 0.4 mM. Incubate 37C with shaking ~ 4 hours (see note).
4) Spike culture with additional antibiotics (another 100ug/ml AMP and 12.5ug/ml TET) and IPTG (to final total concentration
of 0.8mM). Seed plates using culture as is or concentrate cells by centrifugation as per general protocol.
note: the same parameters which give variable results in protein expression using the T7 promoter system are also variable in
this system. Variables to consider: induction temperature (37C vs 30C), induction time (2hr, 4hr, overnight--although
overnight induction never works for me), concentration of IPTG, induction volume, media (LB vs 2xYT or other medias),
additives to induction media (uracil, lactose,etc), etc. Alterations from this "standard" protocol have not significantly or
reproducably improved the RNAi results for the genes we have tested. In addition, “fresh” cells tend to work best. Bacteria
that have been stored on plates at 4C for a long period of time often lose effectiveness: try streaking a new plate from frozen
cells+plasmid or try retransforming HT115(DE3) with the plasmid of interest.
- 公告/新闻




